Processing benchmark results
processing_benchmark_results.RmdMerging benchmark results
The output files from the HPC are organized with one folder per learner and sub-folders for regions. The following snippet merges all the benchmark results using getAllBMRS().
PATH <- "F:/hguillon/research/regional_benchmark/" dirs <- list.dirs(PATH, recursive = FALSE, full.names = FALSE) bmrs <- lapply(dirs, function(dir){getAllBMRS(file.path(PATH, dir), pattern = paste0(dir, "_"))}) bmrs_perf <- lapply(bmrs, function(bmr) bmr$perf) bmrs_tune <- lapply(bmrs[dirs != "baseline"], function(bmr) bmr$tune %>% distinct()) BMR_res <- list() BMR_res$perf <- do.call(rbind, bmrs_perf) BMR_res$tune <- bmrs_tune[[1]] for (i in seq_along(bmrs_tune)[-1]) BMR_res$tune <- full_join(BMR_res$tune, bmrs_tune[[i]])
The associated results in BMR_res are provided with the package.
data("BMR_res") BMR_perf <- getBMR_perf_tune(BMR_res, type = "perf") BMR_perf %>% dplyr::sample_n(10) %>% knitr::kable(digits = 3, format = "html", caption = "benchmark results") %>% kableExtra::kable_styling(bootstrap_options = c("hover", "condensed")) %>% kableExtra::scroll_box(width = "7in", height = "5in")
| task.id | learner.id | iter | multiclass.au1u | acc | mmce | multiclass.aunu | timetrain | FS_NUM |
|---|---|---|---|---|---|---|---|---|
| NC | randomForest | 100 | 0.968 | 0.853 | 0.147 | 0.967 | 130.775 | 45 |
| SECA | naiveBayes | 15 | 0.950 | 0.667 | 0.333 | 0.871 | 0.040 | 11 |
| SAC | naiveBayes | 28 | 0.836 | 0.255 | 0.745 | 0.759 | 0.010 | 5 |
| K | kknn | 19 | 0.944 | 0.848 | 0.152 | 0.939 | 0.000 | 6 |
| SC | nnTrain | 54 | 0.819 | 0.462 | 0.538 | 0.784 | 1205.046 | 6 |
| SJT | kknn | 38 | 0.992 | 0.750 | 0.250 | 0.992 | 0.010 | 8 |
| K | naiveBayes | 18 | 0.904 | 0.656 | 0.344 | 0.905 | 0.010 | 10 |
| NC | randomForest | 21 | 0.939 | 0.794 | 0.206 | 0.941 | 64.493 | 15 |
| SFE | featureless | 50 | 0.500 | 0.095 | 0.905 | 0.500 | 0.030 | 30 |
| SC | kknn | 88 | 0.858 | 0.615 | 0.385 | 0.849 | 0.030 | 18 |
Processing benchmark results
Performance plots
makeAverageAUCPlot(BMR_perf) + ggplot2::scale_x_continuous(breaks = c(5, 25, 50))

makeAverageAccPlot(BMR_perf) + ggplot2::scale_x_continuous(breaks = c(5, 25, 50))

makeTotalTimetrainPlot(BMR_perf) + ggplot2::scale_x_continuous(breaks = c(5, 25, 50))

Model selection
The model selection is handled by examining statistical differences between the distribution of performance metrics for successive iterations of model training with an increasing number of predictors. On the following graph, the brackets indicate statistically significant differences between the distribution assessed by a Dunn test for the SAC region and the randomForest learner.
makeExampleModelSelectionPlot(BMR_perf, lrn = c("randomForest"), window_size = 7)[["SAC"]]
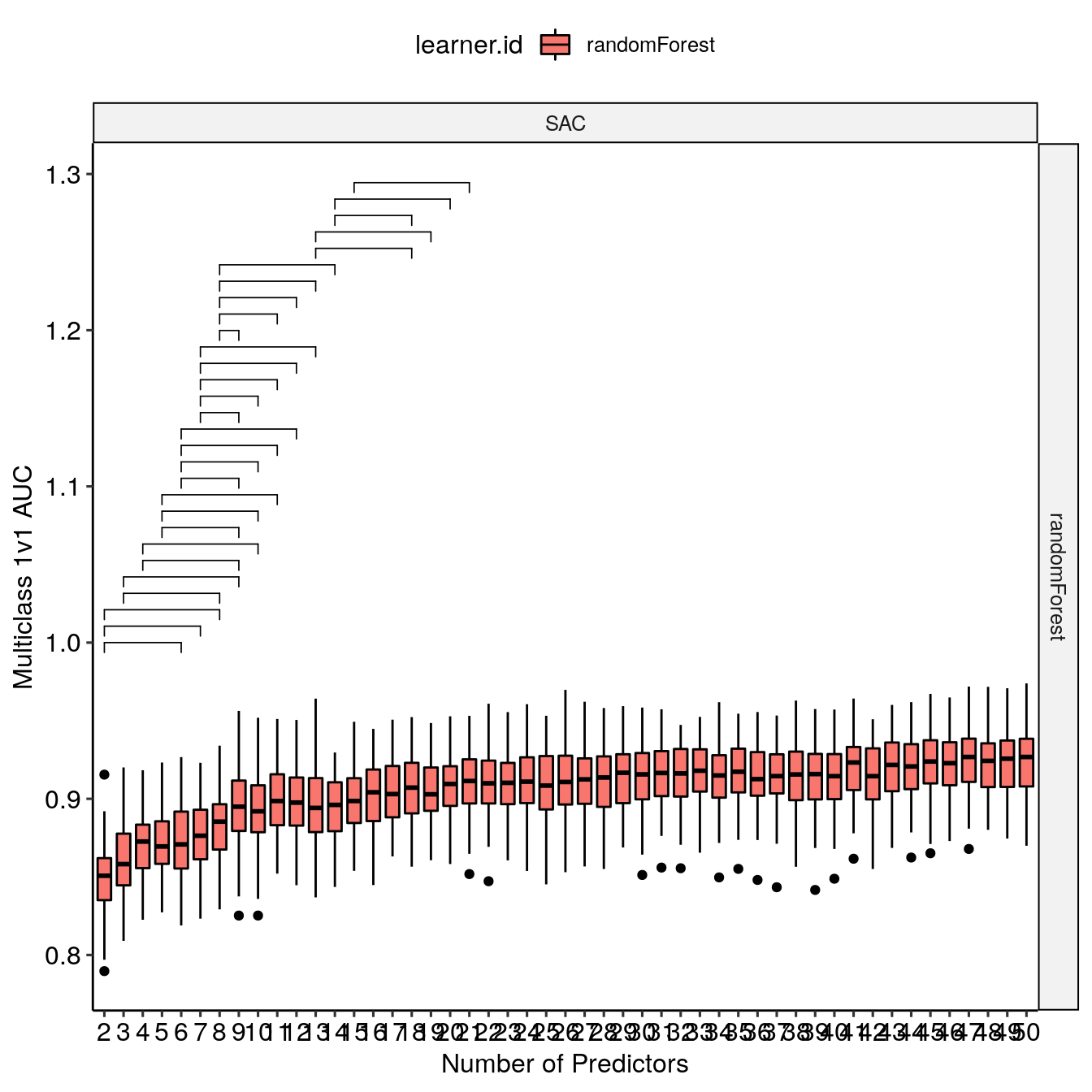
The p-value is internally ajusted to take into account multiple comparison with a Bonferroni correction. The search window influence (window_size) is examined with makeWindowInfluencePlot() which requires use to declare where we are storing the names of the selected features for each iterations.
selectedPATH <- system.file("extdata/selectedFeatures/", package = "RiverML")
makeWindowInfluencePlot(BMR_perf, selectedPATH, lrn = "randomForest") + ggplot2::xlim(c(0,20))

The window size selection is mainly based on the evolution of the number of optimal predictors per classes in a given region of study with respect to the window size. This balances a trade-off between two effects. A large window increases the likelihood to find a statistically different performance distribution is high and selects a large number of predictors. A small window decreases the likelihood to find statistically significant difference but might miss the inclusion of an important predictor leading to a significant increase in performance. In our example above, with a window size of 8, there is a major jump in the number of predictors per class which increases the likelihood of overfit for the optimal models.
Feature importance
The optimal feature sets are obtained for each learner by using get_bestFeatureSets().
bestFeatureSets <- get_bestFeatureSets(BMR_perf, selectedPATH, window_size = 7)
A convenience function, makeAllFeatureImportancePlotsFS(), creates all the feature importance plots from the standpoint of feature selection, that is evaluating which feature is present in the optimal feature sets. Here is an example for the SFE region.
FeatureImportancePlots <- makeAllFeatureImportancePlotFS(BMR_perf, selectedPATH, bestFeatureSets) FeatureImportancePlots[["SFE"]]
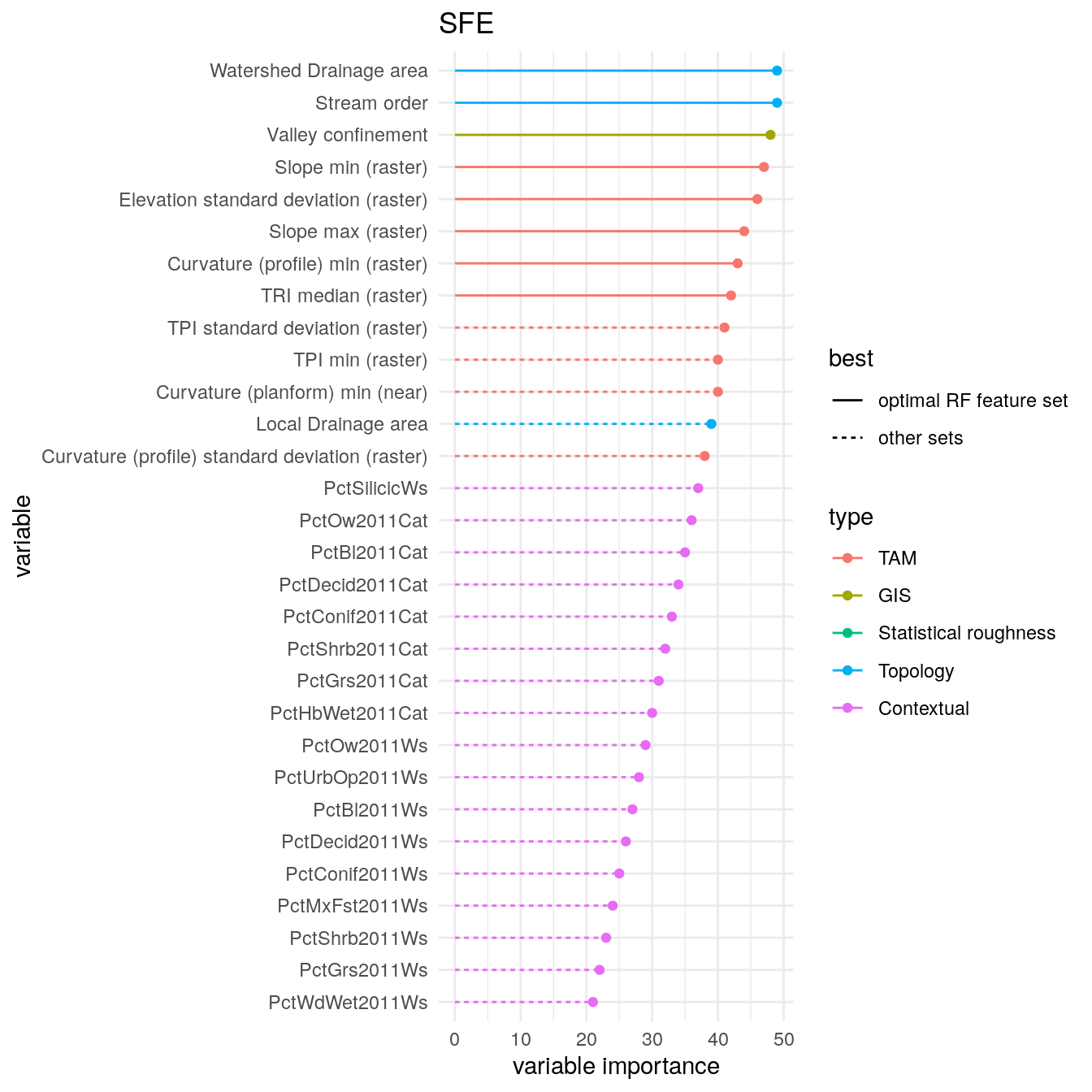
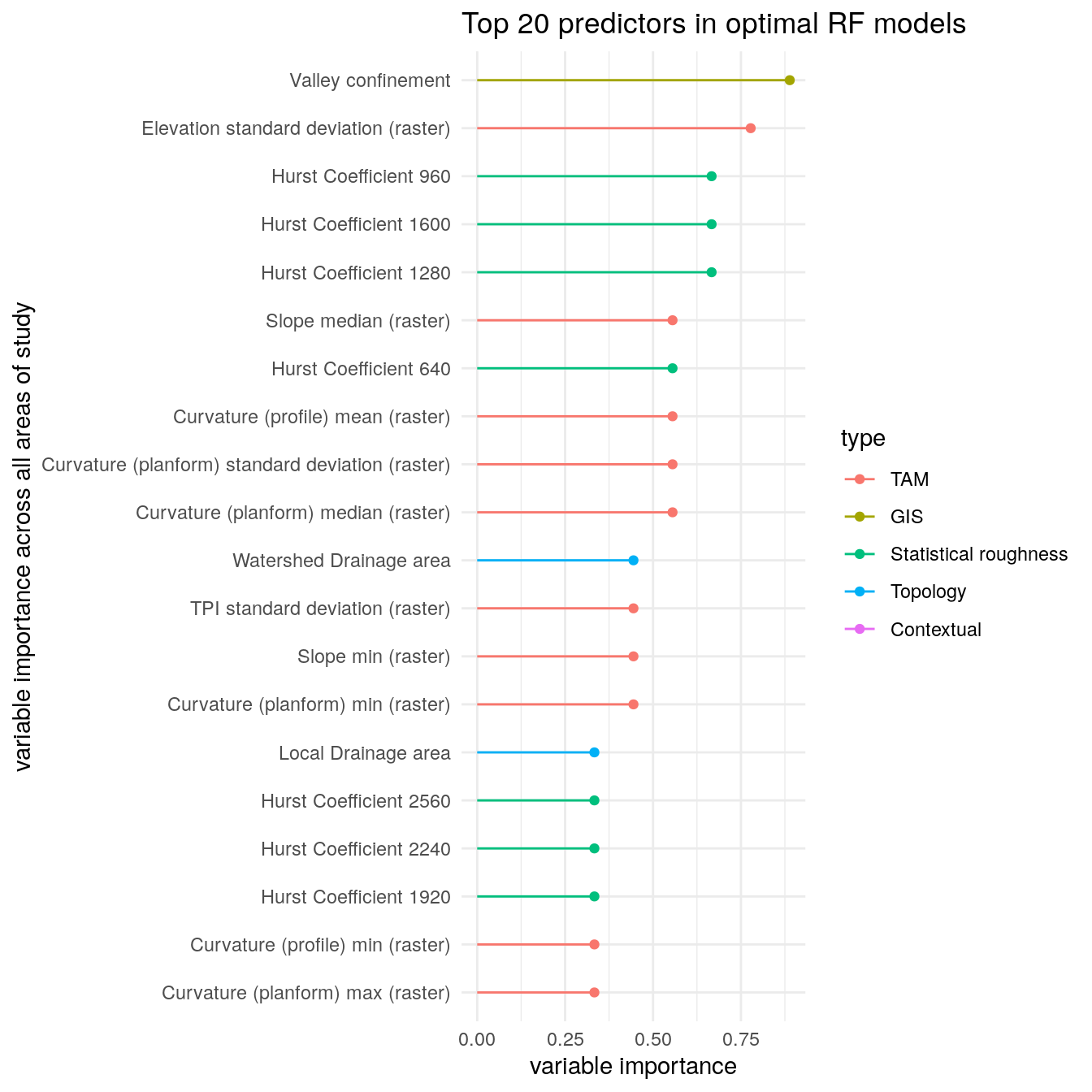
We can also investigate which features are often selected across all regions with getFreqBestFeatureSets().
freq_bestFeatureSets <- getFreqBestFeatureSets(bestFeatureSets) freq_bestFeatureSets %>% dplyr::rename(var = group, Overall = prop) %>% dplyr::arrange(-Overall) %>% makeFeatureImportancePlot(first = 20) + ggplot2::labs(title = "Top 20 predictors in optimal RF models", y = "variable importance across all areas of study")
Performance of optimal models
We now turn to analyzing the tuning result contained in BMR_tune.
BMR_tune <- getBMR_perf_tune(BMR_res, type = "tune") BMR_tune %>% dplyr::sample_n(10) %>% knitr::kable(digits = 3, format = "html", caption = "benchmark results") %>% kableExtra::kable_styling(bootstrap_options = c("hover", "condensed")) %>% kableExtra::scroll_box(width = "7in", height = "5in")
| task.id | learner.id | iter | max.number.of.layers | hidden | activationfun | output | numepochs | learningrate | batchsize | momentum | hidden_dropout | visible_dropout | multiclass.au1u | acc | mmce | multiclass.aunu | timetrain | FS_NUM | mtry | cost | kernel |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SFE | svm | 22 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.963 | 0.841 | 0.159 | 0.959 | 0.085 | 41 | NA | 0.25 | linear |
| SC | svm | 65 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.928 | 0.764 | 0.236 | 0.910 | 0.037 | 18 | NA | 4.00 | linear |
| NCC | svm | 7 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.954 | 0.773 | 0.227 | 0.943 | 0.116 | 42 | NA | 1024.00 | linear |
| SC | randomForest | 4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.866 | 0.592 | 0.408 | 0.819 | 0.083 | 3 | 3 | NA | NA |
| NCC | randomForest | 61 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.956 | 0.732 | 0.268 | 0.939 | 0.300 | 25 | 6 | NA | NA |
| SCC | randomForest | 35 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.934 | 0.669 | 0.331 | 0.916 | 0.142 | 4 | 2 | NA | NA |
| SECA | randomForest | 34 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.954 | 0.753 | 0.247 | 0.936 | 0.104 | 18 | 6 | NA | NA |
| SCC | svm | 49 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.918 | 0.568 | 0.432 | 0.873 | 1.000 | 11 | NA | 256.00 | linear |
| SFE | nnTrain | 80 | 2 | 30 | tanh | softmax | 20 | 0.05 | 16 | 0.8 | 0 | 0 | 0.969 | 0.812 | 0.188 | 0.960 | 0.806 | 18 | NA | NA | NA |
| SECA | svm | 9 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.902 | 0.470 | 0.530 | 0.830 | 0.096 | 3 | NA | 512.00 | linear |
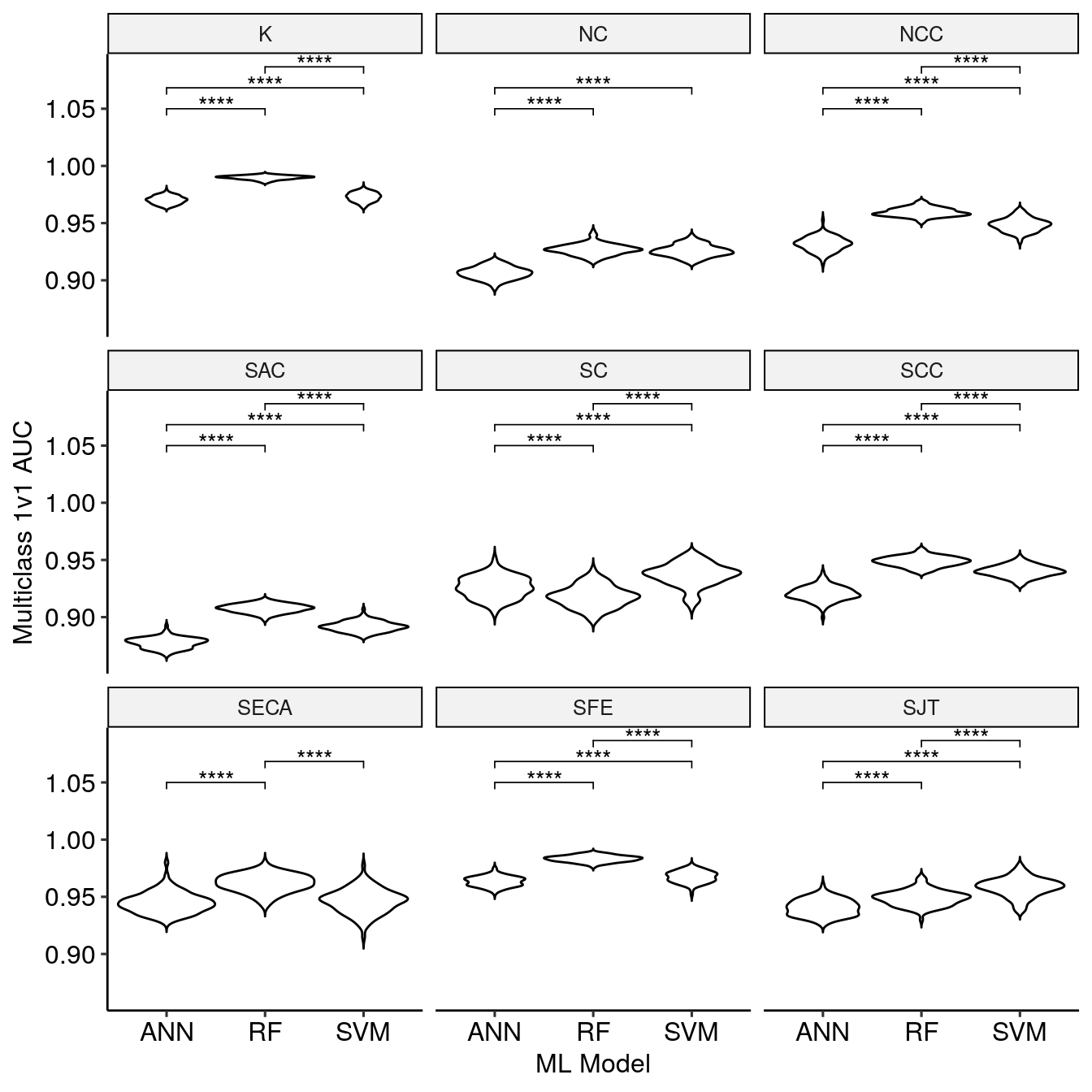
This data is subsetted by the optimal model selection with getBestBMRTune(). The resulting data.frame can be visualized with makeBestTuneAUCPlot() to compare the results of the optimal models.
bestBMR_tune <- getBestBMRTune(BMR_tune, bestFeatureSets) makeBestTuneAUCPlot(bestBMR_tune)
Tuning entropy
Because the benchmark used nested resampling, we have access to a distribution of best tune hyper-parameters which can be visualized with getTunePlot().
getTunePlot(bestBMR_tune, "nnTrain")
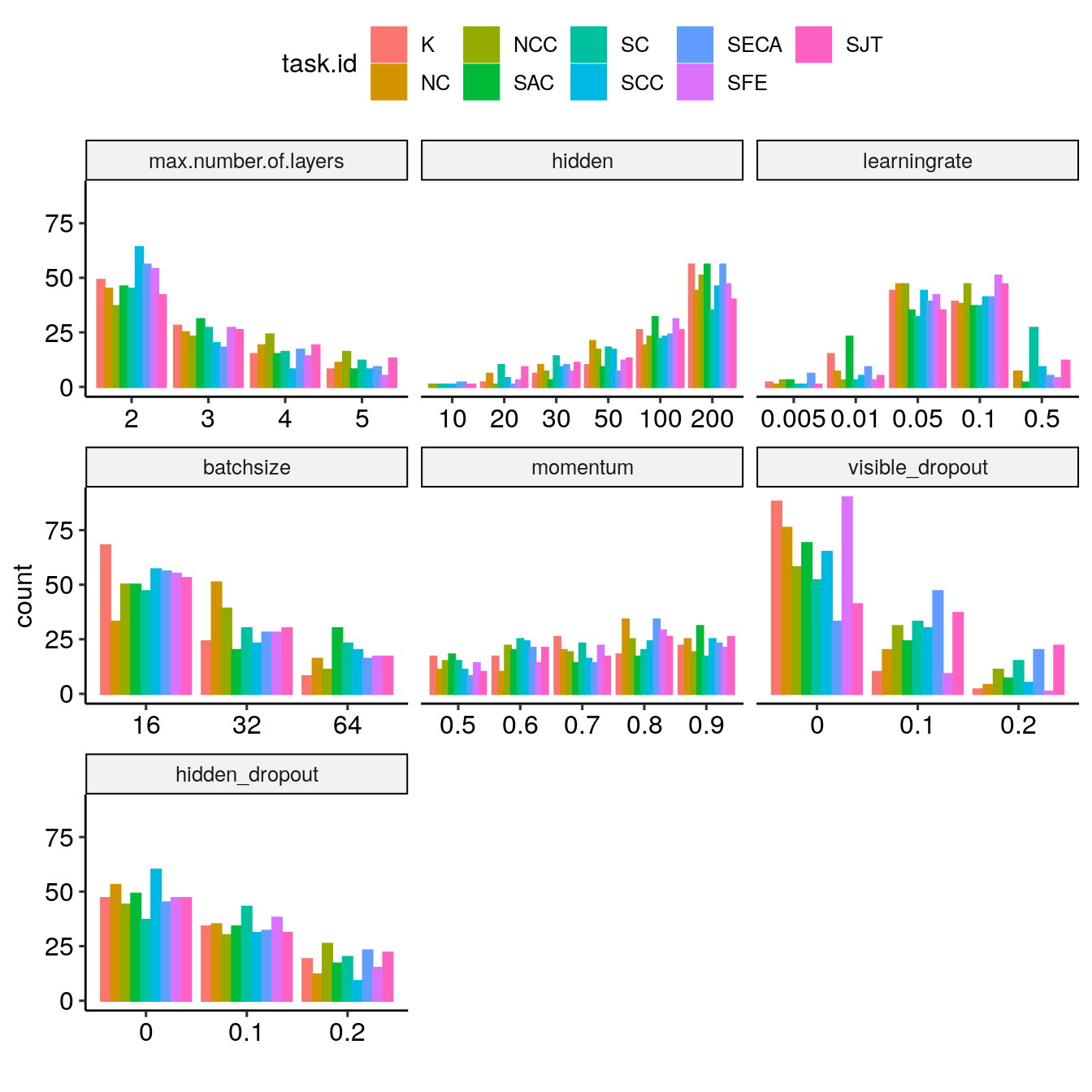
In addition, while the previous plot focused on the optimal, we can investigate how the tuning entropy evolves with the number of selected features using getBMRTuningEntropy(), getBestBMRTuningEntropy(), and makeTuningEntropyPlot(). Here is one example for the SFE region.
TUNELENGTH <- 16 BMR_lrnH <- getBMRTuningEntropy(BMR_tune, TUNELENGTH) bestBMR_lrnH <- getBestBMRTuningEntropy(BMR_lrnH, bestFeatureSets, bestBMR_tune) p_Hnorms <- makeTuningEntropyPlot(BMR_lrnH, bestBMR_lrnH) p_Hnorms[["SFE"]]
